For Jupyter Users
1. g2nb Meta-Package
g2nb consists of a collection of Python packages and extensions for JupyterLab. It provides a variety of tools for interacting with powerful analysis platforms from within a Jupyter environment. These tools include:
- Visual widgets for running bioinformatic analyses and submitting these analyses to a GenePattern or Galaxy server and for starting interactive visualizations using IGV and Cytoscape.
- Tools for visually sending results to downstream steps in a bioinformatic workflow.
- A WYSIWYG-style rich text editor for Markdown cells.
- Python libraries for programmatically working with the results of the analyses.
g2nb usage is free and the software is open source under the BSD license.
2. What is GenePattern?
GenePattern is a powerful platform for reproducible bioinformatics research. It provides hundreds of analytical tools for the analysis of gene expression (RNA-seq and microarray), sequence variation and copy number, proteomic, flow cytometry, and network analysis.
For more information visit the GenePattern website. For introductory information about GenePattern, please see our 10-minute tutorial and/or check out one of our video tutorials.
3. What is Galaxy?
Galaxy is an open source, web-based platform for data intensive biomedical research. You can install your own Galaxy instance or use the main Galaxy server and choose from thousands of tools from the Tool Shed.
For more information visit the Galaxy tutorial page.
4. What is Cytoscape?
Cytoscape is an open source software platform for visualizing complex networks and integrating these with any type of attribute data. A lot of Apps are available for various kinds of problem domains, including bioinformatics, social network analysis, and semantic web.
For more information visit the Cytoscape website.
5. What is IGV?
The Integrative Genomics Viewer (IGV) is a high-performance, easy-to-use, interactive tool for the visual exploration of genomic data. It supports flexible integration of all the common types of genomic data and metadata, investigator-generated or publicly available, loaded from local or cloud sources.
For more information visit the IGV webpage.
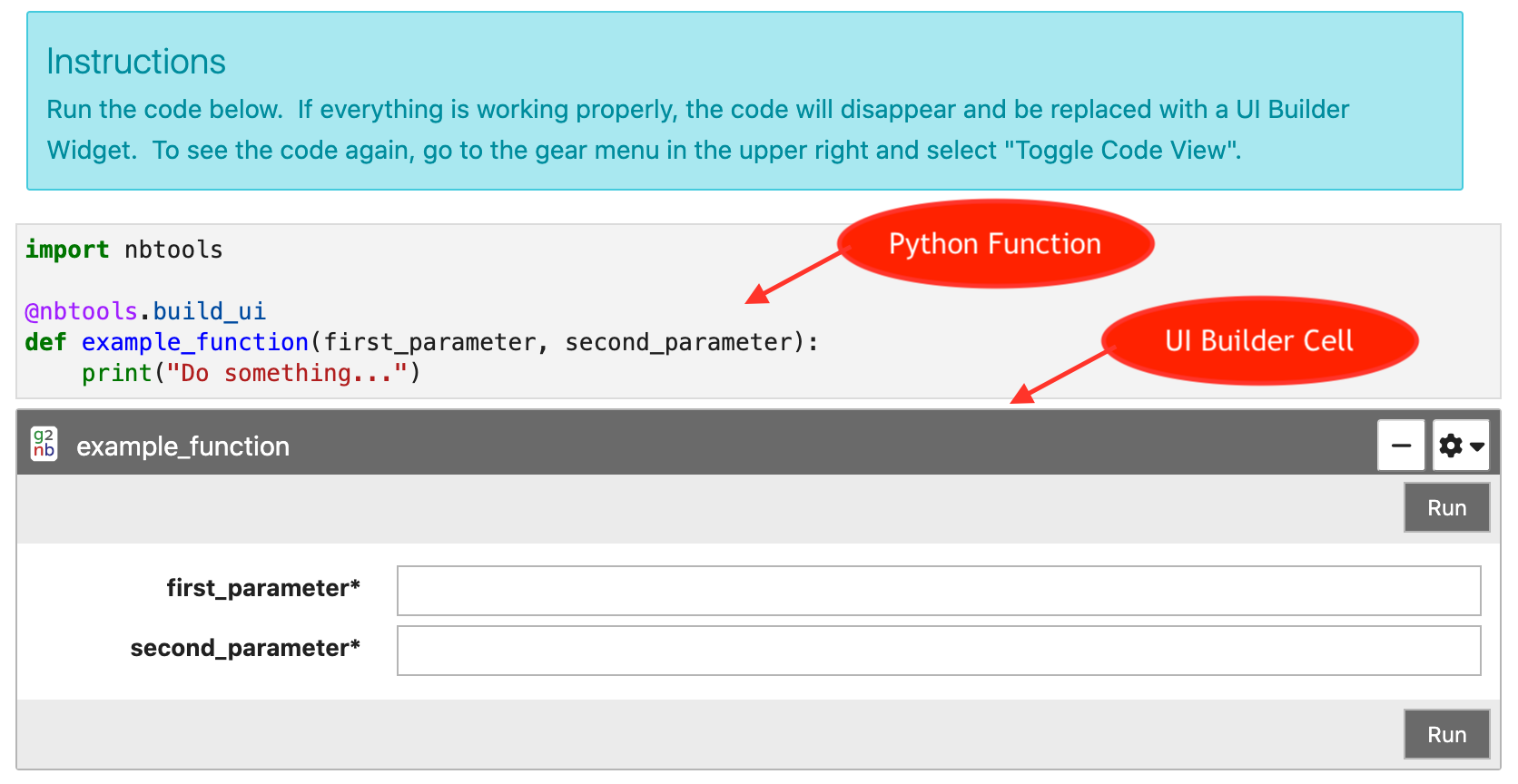
7. UI Builder
The UI Builder is a way to display any Python function or method call as
an interactive widget. This will render the parameters of the function
as a web form which can be filled out with numbers, string literals or
with references to existing Python variables. Clicking the Run button on
the widget will validate the form and execute the function using the
supplied values.
The UI Builder will use any existing docstring for the function as a description in the widget, will infer parameter types from default values and will display parameter annotations as helpful text near each input.
To use render a function, simply import the UIBuilder class from the
nbtools package and pass the function to its constructor.
For more information, see the UI Builder documentation